Every now and then I work with new groups as they embark on a journey incorporating peptides and peptide synthesis into their research. More often than not, no one in the group has experience with peptide synthesis as they are just getting operations off the ground. As a result, one of the most common questions I receive from these groups is how much amino acid should be used during synthesis.
In today’s post though I will address the number of equivalents of amino acid. Large numbers of amino acid equivalents can often be used to drive coupling reactions to near completion, but the question today is how few equivalents can be used to successfully synthesize your peptide.
This becomes an important question when you start thinking about the cost of peptide synthesis. The price of many amino acids has come down significantly over the years, but there are still some amino acids that are exorbitantly expensive (think isotopically labeled amino acids). In order to minimize the overall cost of a synthetic peptide, it is critical to ensure that the peptide is successfully synthesized in a single attempt. In a previous post, I discussed the importance of amino acid concentration particularly for synthesis of long peptides, but the question remains, how many equivalents of amino acid should be contained in that liquid volume.
To address this question, I chose to synthesize Helix A, an 18 amino acid amphipathic helical peptide. This peptide is of reasonable length yet contains evenly dispersed hydrophobic and hydrophilic amino acids making synthesis relatively straightforward. I synthesized the peptide three times, varying the number of amino acid equivalents, but keeping the scale, coupling reagents and reaction temperature and time consistent.
My goal was to identify a minimal number of amino acid equivalents that can be used to successfully synthesize a peptide without significant reaction optimization. You could certainly use an extreme minimum number of equivalents, but a successful synthesis will require fairly extensive coupling reaction optimization.
So to start, I did the thing that no peptide chemist wants to do. I prepared Helix A using 1 equivalent of amino acid. Using my Initiator+ Alstra™ peptide synthesizer, I chose DIC and oxyma as my coupling reagents and each amino acid was allowed to react for 5 minutes at 75°C. As many of you would expect, this synthesis totally failed, Figure 1.
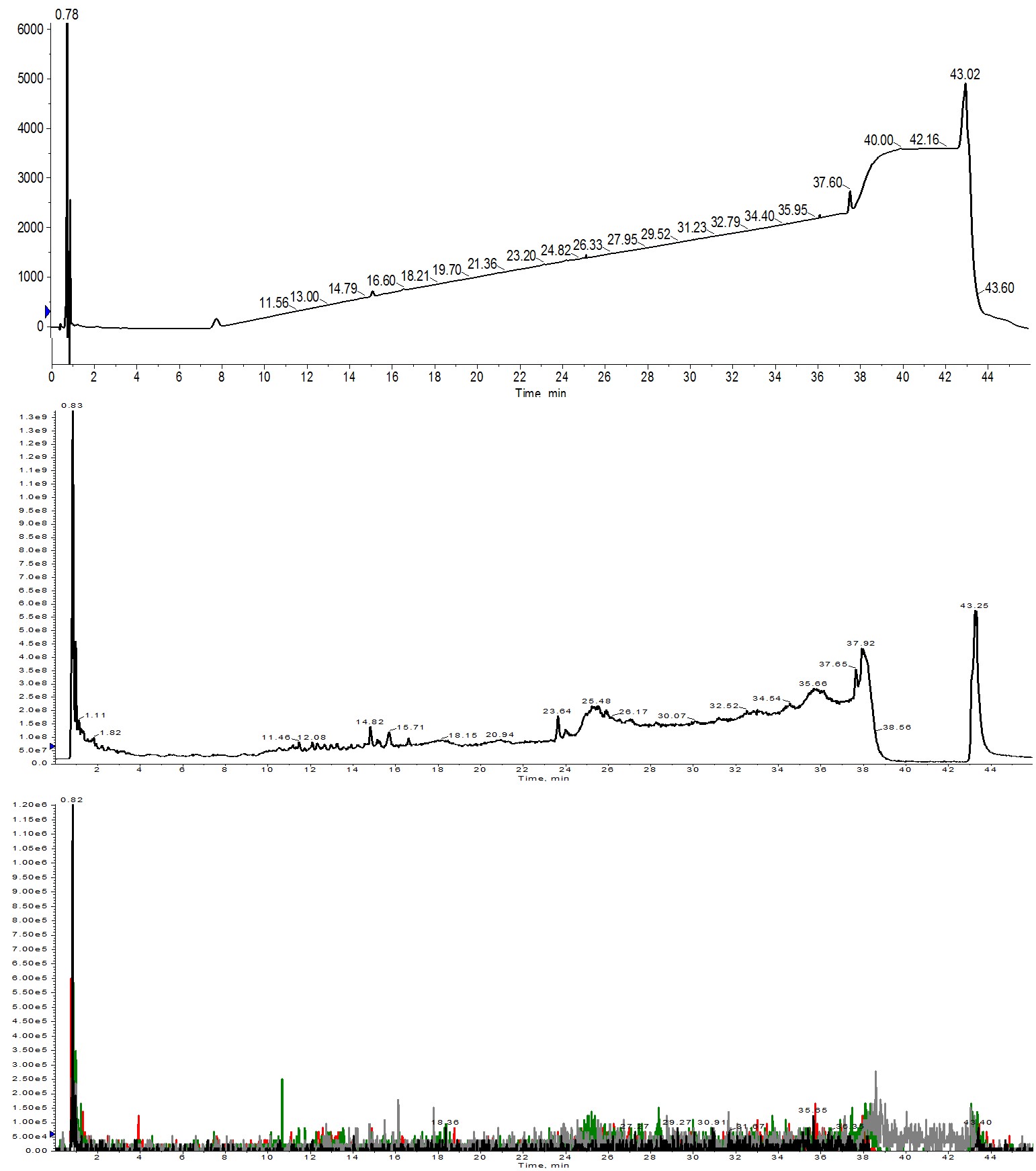
Figure 1: Crude analytical HPLC-MS chromatography for Helix A synthesized using 1 equivalent of amino acid. No peptide was identified in the UV absorbance trace (top), the total ion count (middle) or even when search for specific ions within the TIC (bottom). Extracted ions evaluated were 1113 (m+2, black), 742 (m+3, red), 557 (m+4, green) and 445 (m+5, grey).
There are no peaks in the UV and very few peaks even in the mass TIC. When I attempted an ion extraction, the desired peptide charge states could not be identified. There are most likely a couple of things at play here. First and foremost is the fact that the amino acids are used in equimolar quantities relative to the resin functionalization. This demands that every single amino acid molecule be successfully activated by the coupling reagents and then consumed in the coupling reaction for the synthesis to be successful. The probability of this actually occurring is small. As second factor to consider is concentration. Both the amino acid and DIC solutions were diluted by half to 0.1 M in order to achieve the minimal liquid volume in the reactor vial for efficient and even heating. Diluting the samples further decreases the probability that the amino acid will productively interact with the coupling reagents in solution and then the activated amino acid interacting with the amino terminus of the growing peptide chain. This result highlights why we use superstoichiometric equivalents of amino acid relative to the resin loading.
In the second synthesis, I increased the number of amino acid equivalents to 2 and brought the amino acid and DIC concentration up to their normal 0.2 M concentration. This synthesis did yield peptide product, although not the desired peptide product, Figure 2.
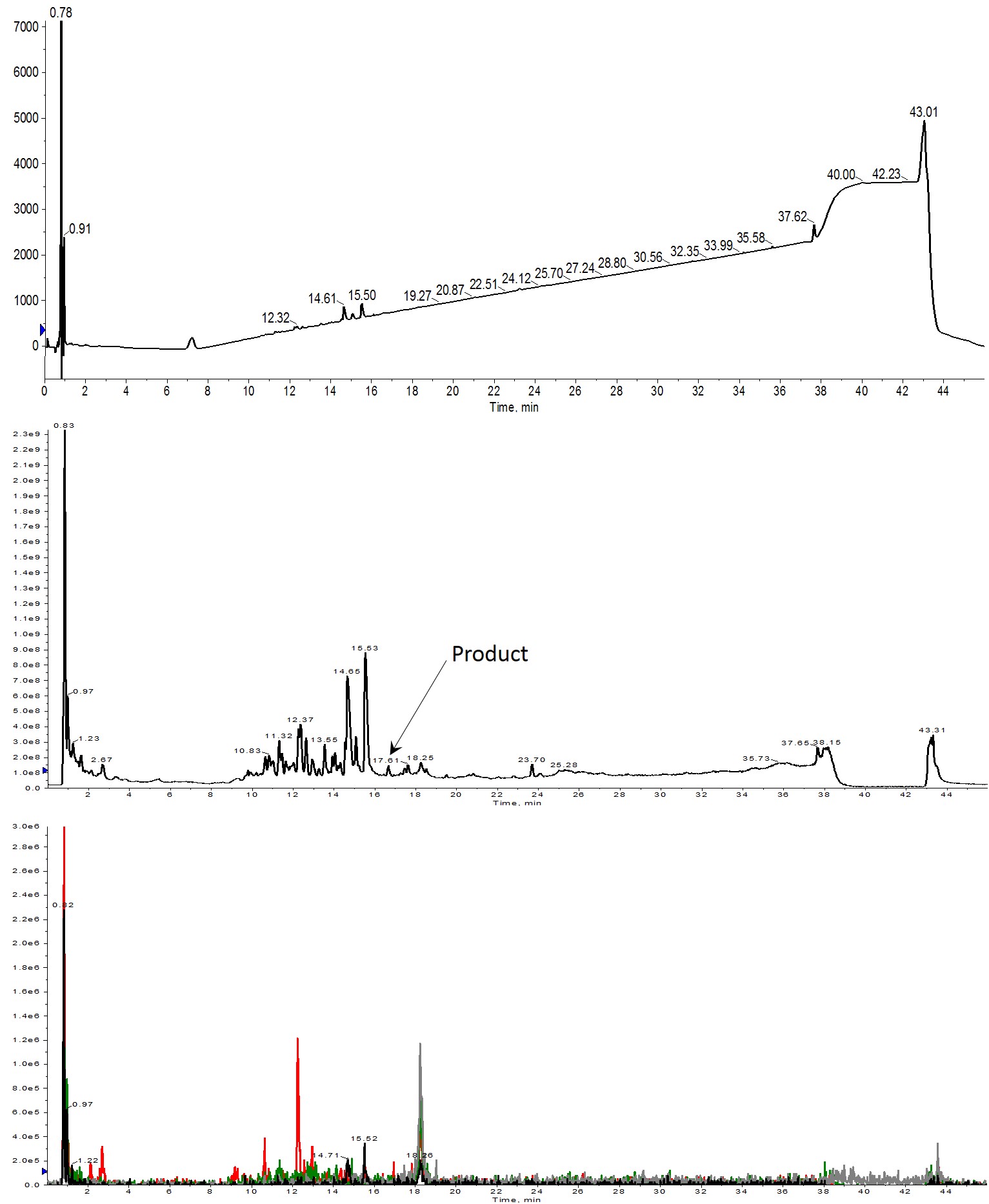
Figure 2: Crude analytical HPLC-MS chromatograms of Helix A synthesized using 2 equivalents of amino acid. No peptide product is observed in the UV spectrum (top). Very small amounts are evident in the total ion count (middle) and which is further verified with the extracted ion chromatogram (bottom). Extracted ions evaluated were 1113 (m+2, black), 742 (m+3, red), 557 (m+4, green) and 445 (m+5, grey).
Increasing the number of equivalents did provide a modest improvement as you would expect. Unfortunately, very little of the desired peptide was identified in the solution, Figure 2, bottom. I was able to identify several masses that correspond to peptides with 7, 9, and 13 amino acids (14.65 and 15.53 min retention time respectively), giving me hope that the next increase could yield my desired full length peptide.
For my third synthesis, I increased my amino acid equivalents to 3, keeping the concentration at 0.2M just like the previous synthesis. Finally, I was able to identify my desired peptide product in the mass spectrum and even more satisfying is the fact that the desired peptide is in fact the majority product by UV, Figure 3.
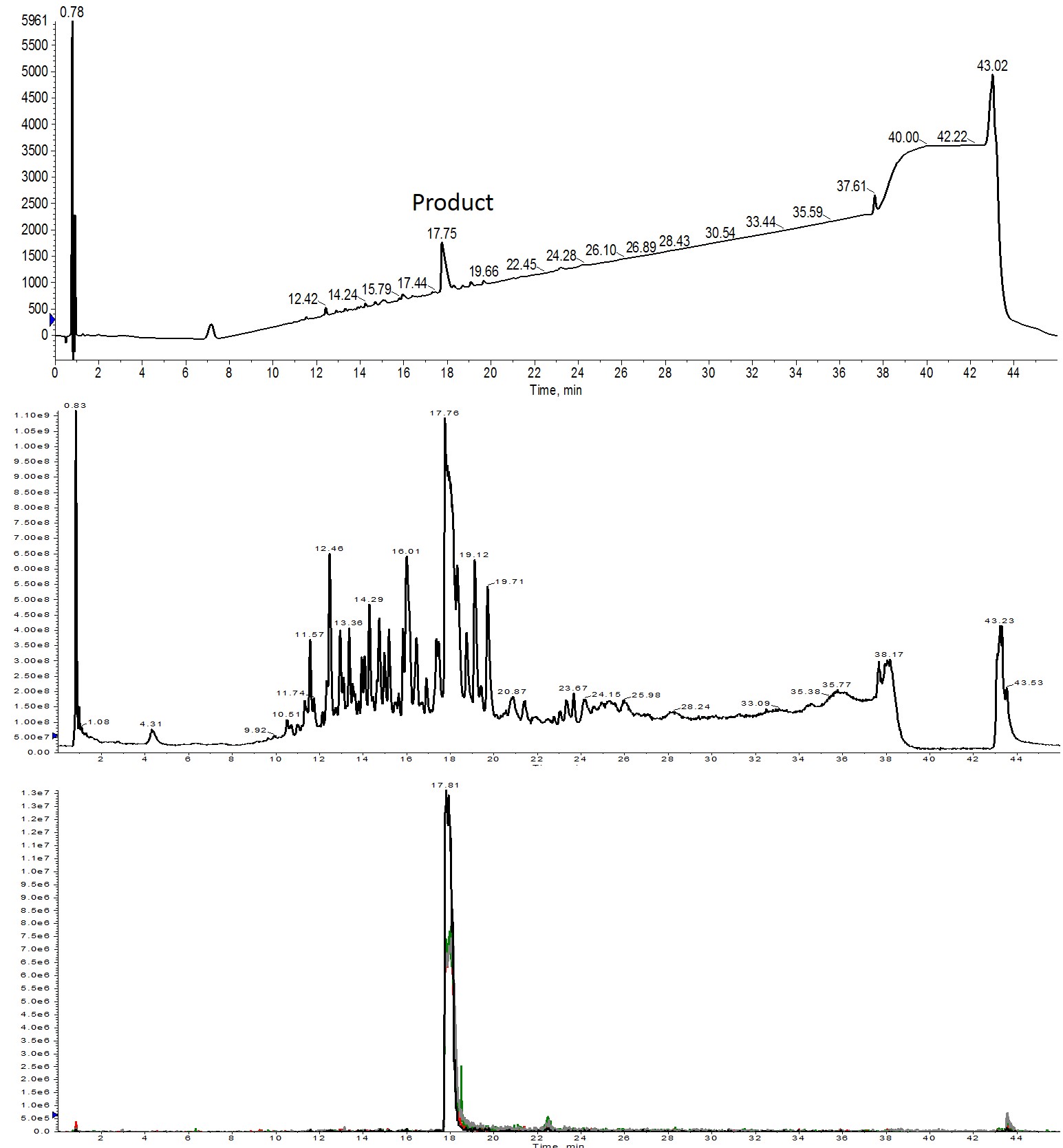
Figure 3: Crude analytical HPLC-MS analysis of Helix A synthesized using 3 equivalents of amino acid. The desired product is present as the main peak in the UV spectrum (top) and is also the most abundant ion present in the total ion count spectrum (middle). All four charge states are abundant and overlap when extracted from the total ion chromatogram (bottom). Extracted ions evaluated were 1113 (m+2, black), 742 (m+3, red), 557 (m+4, green) and 445 (m+5, grey).
The UV trace suggests that purification of this peptide will be relatively straightforward, further supporting the observation that 3 equivalents of amino acid are necessary to ensure high yielding, relatively clean crude peptide product without the need for significant coupling reaction optimization.
While you could certainly improve the overall yield and potentially increase the overall purity of the sample by increasing the number of equivalents, you can certainly reduce your overall cost of synthesis by reducing the number of equivalents as I have demonstrated above. Please remember this is a peptide of somewhat average length, these conclusion will likely not hold for peptides of significant length.
To learn more about how Biotage tools can be used in synthesis of complex peptides, follow the link below to see my webinar on Synthesizing Disulfide-rich Peptides:

 Organic Workflow
Organic Workflow Peptide Workflow
Peptide Workflow Scale-Up Flash Purification
Scale-Up Flash Purification  Sample Preparation
Sample Preparation Biomolecule Purification
Biomolecule Purification Oligo synthesis
Oligo synthesis Scavengers and Reagents
Scavengers and Reagents Service & Support
Service & Support Accessories & Spare parts
Accessories & Spare parts Investors
Investors Reports & News
Reports & News The Share
The Share Corporate Governance
Corporate Governance Calendar
Calendar Sustainability
Sustainability Our Offering
Our Offering Our History
Our History Our Locations
Our Locations Leadership
Leadership